Documents in the health domain show specific vocabulary and linguistic structure. If you take a look at clinical Records or Electronic Health Records (EHR), you will see that it is also made up of unstructured data (that is, free text). This free text contains weird names of drugs and diseases that are even difficult to read. For all these reasons, text analytics techniques must be adapted to the health domain.
We have put together a number of resources in a demo that shows how MeaningCloud can tag drug names, symptoms, diseases, procedures, and so on.
See the free demo: https://www.meaningcloud.com/demos/health-text-analytics-demo
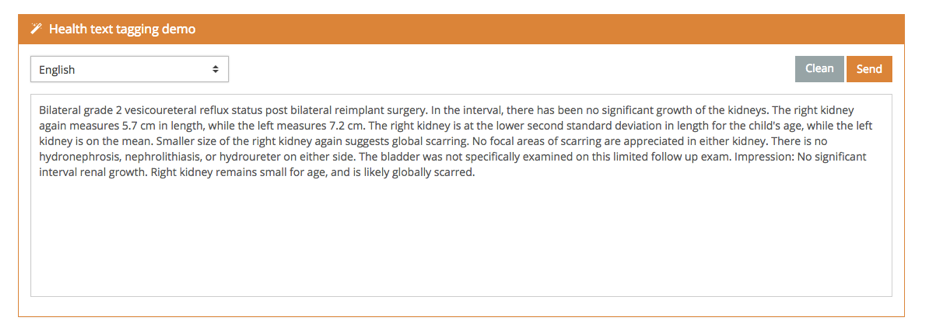
The demo also shows how we have linked these mentions with sources of external information.
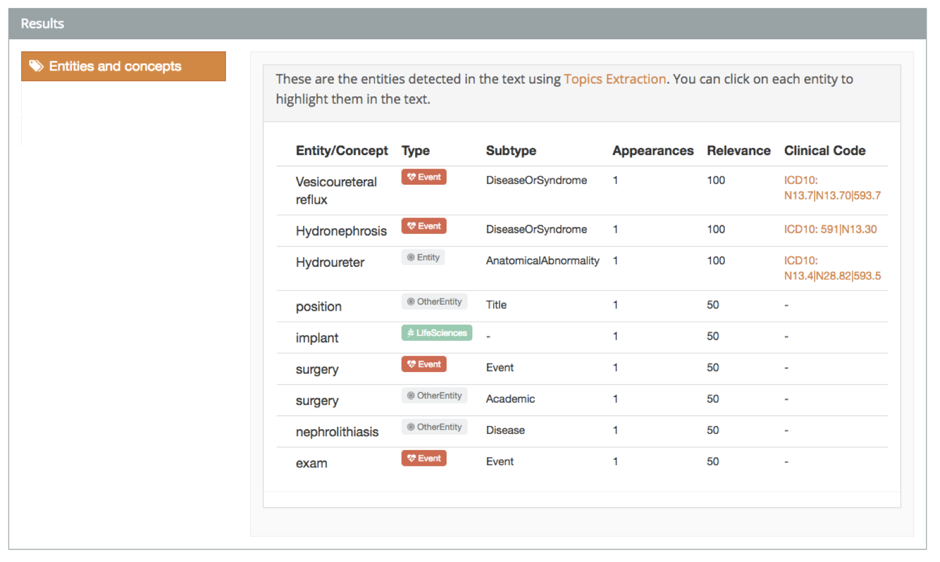
With this demo, you can tag texts both in Spanish and English. However, the full version supports Spanish, English, French, Italian, Portuguese, and Catalan.
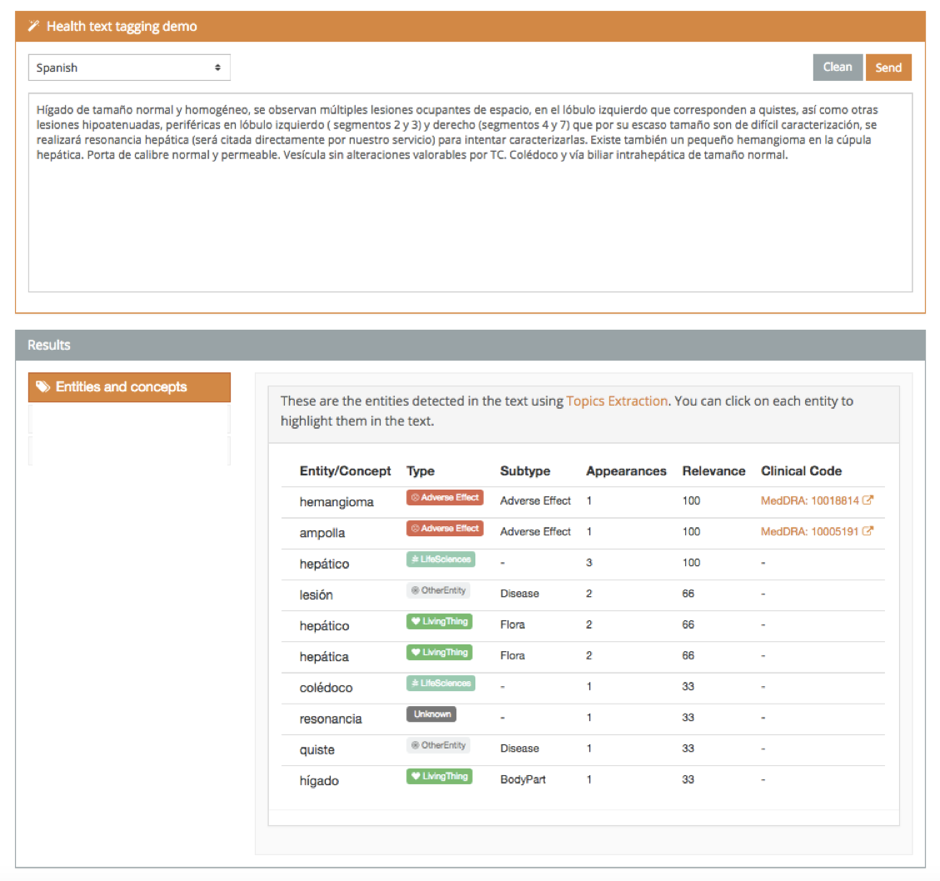
Adverse drug reactions
It is well-known that adverse drug reactions (ADRs) are a prominent health matter, being the fourth cause of demise in hospitalized patients [1]. Thus, the field of pharmacovigilance has received lots of attention due to the growing impact of drug safety events [2] as well as their high associated costs [3]. This demo detects and tags Adverse Effects.
We have now a dedicated business exclusively focused on the health and pharmaceutical sectors
Konplik.Health begins operations with the health-related assets from MeaningCloud, including its leading natural language processing, deep semantic analysis, AI platform, and adaptations specific to the life sciences.
How does it work?
MeaningCloud leverages several Semantic APIs in SaaS (Software as a Service) mode to extract elements of meaning (topics, facts, opinions, relationships, etc.) from all kinds of unstructured multimedia content.
1. Morphosyntactic Parsing
Text analytics is performed by the MeaningCloud Lemmatization, PoS and Parsing API which follows a dictionary-based approach to morphosyntactical analysis.
This step is of great importance to the disambiguation step that comes afterward, due to the great ambiguity that exists in medical texts.
The currently supported languages are Spanish, English, French, Italian, Portuguese, and Catalan.
2. Topic Analyzer
Several health-related dictionaries were created to perform Named Entity Recognition (drugs, diseases, ADRs, and others) and integrated into the MeaningCloud Topic Extraction API.
The currently supported languages are Spanish, English, French, Italian, Portuguese, and Catalan.
3. Medical Events Filter
It filters all the entities that have been annotated by the Topic Analyzer and which are not from the medical domain. Only drug, effect, and disease entities are kept in the system.
4. Disambiguation
A set of rules that uses linguistic features like the morphosyntactic information provided by the parser, together with co-occurrence information on drugs and diseases are used to filter out terms that are not likely to be mentions of medical events.
Resources: Drugs, Diseases, and Effects
There are several semantic resources integrated into the system each of them intended to detect a different type of named entity or relation as explained below.
MedDRA
MedDRA is the adverse event classification dictionary approved by the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH), and therefore a very reliable resource for the adverse events.
MedDRA supports ten languages and is composed of a five-level hierarchy which goes from most general to most specific. The two lower levels from MedDRA PT (Preferred Terms) and LLT (Lowest Level Terms) were extracted to implement the adrsMedDRA dictionary for ADRs detection. Each LLT is a single medical concept for a symptom, sign, disease diagnosis, therapeutic indication, investigation, surgical or medical procedure, and medical, social, or family history characteristic.
Finally, the information we obtained from this resource is: 13,245 PT adverse effects and 35,259 LLT adverse effects.
UMLS-SNOMED CT
UMLS, developed by the National Library of Medicine (NLM), is a comprehensive list of medical terms mainly focused on developing computer systems suitable for understanding the specific vocabulary which is normally used in biomedicine and health care literature. One of the resources integrated into UMLS is SNOMED CT (Systematized Nomenclature of Medicine – Clinical Terms), a glossary, accessible in Spanish, that consists of concepts, descriptions, and relationships to represent information and clinical knowledge. UMLS is structured in several semantic categories (substances, organisms, health care activity, etc.). Three of these categories (‘Diseases or Syndromes’, ‘Mental or Behavioral Dysfunction’, and ‘Neoplastic process’) have been chosen in order to create the dictionary for diseases and symptoms.
Some of the extracted terms were PT and other synonyms. The PTs were set as canonical expressions in the dictionary, and their synonyms were considered aliases.
An extra information field in the entries is the UMLS CUI (Concept Unique Identifier), a code which relates a specific medical term to a set of resources included in UMLS. As a matter of fact, some terms are included in both the disease dictionary and the adverse effect one.
The information we obtained from UMLS Database is 42,548 main diseases and 23,677 diseases synonyms.
CIMA
CIMA is a resource provided and maintained by The Spanish Agency for Medications and Healthcare Products (AEMPS). It is an application which includes all authorized drugs in Spain. The application contains the following information on authorized drugs in Spain: the name, active substance(s), marketing authorization holder’s name, national code, data sheet, package insert, authorization date, ATC (Anatomical, Therapeutic Chemical classification system) code, and others. The drug’s data sheet includes the drug’s description, indications, dosage, precautions, and contraindications, adverse reactions, pharmaceutical information and properties. The package insert is the document found inside the box of the medication to inform the patient of the data sheet details.
From CIMA files, 16,418 drugs, 2,228 active substances, and 3,659 brand drugs were obtained. Additionally, 4,817 drug-related terms were obtained from Vademecum (a guide of pharmaceutical products that includes over 18,200 drugs) and from MedlinePlus, the National Institutes of Health’s (NIH) website for patients. These terms compose the gazetteer DrugsGaz.
ATC SYSTEM
In order to discover relationships between brand names and active substances, we use the ATC system which consists of a set of alphanumeric codes developed by the WHO to classify drugs and other medical products organized in 5 levels (see Figure 3 below). Level 1 represents the part of the body where the drug performs its activity; level 2 represents the therapeutic group; level 3 is about the pharmacological group of the drug; level 4 is the chemical group; and level 5 concerns the active substance group. Therefore, the ATC system is the key to discovering relationships between drugs and brand names. Wikipedia has a complete, well-structured article dealing with the ATC codes in Spanish that has been crawled to obtain all the existing Spanish ATC codes (4,361 in all).
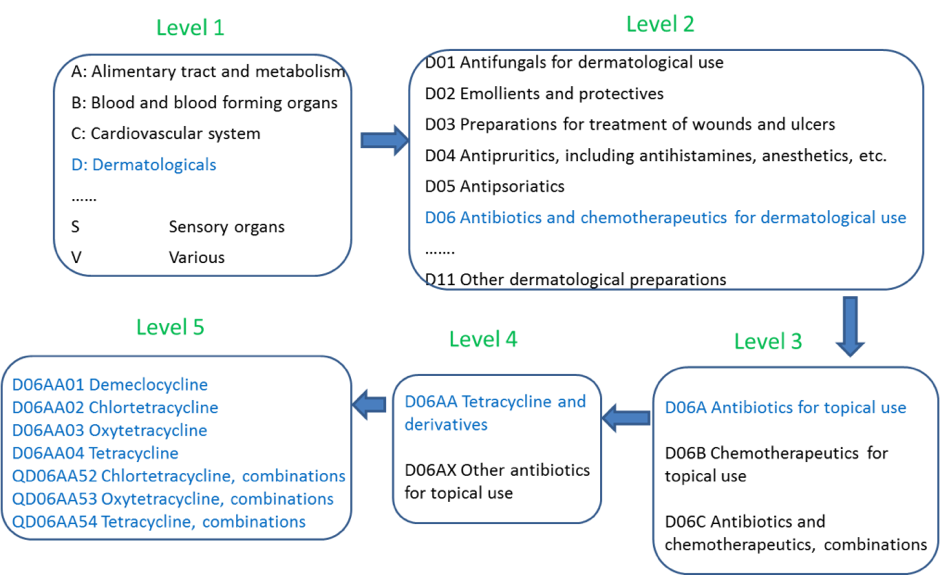
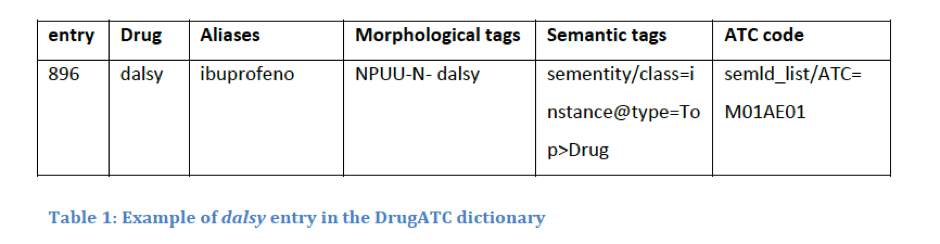
Thanks to the information on drugs that the ATC provides (therapeutic and chemical characteristics), the system is able to relate the drugs and is able to categorize them by active substance, chemical group, or pharmacological group. This is possible due to the classification hierarchy. ATC codes are split into five levels. For example, the ATC code M01AE01 is divided into: Anatomical main group (M), therapeutic main group (01), therapeutic/pharmacological subgroup (A), chemical/therapeutic/pharmacological subgroup (E) and the chemical substance (01), forming the final M01AE01.
The final M01AE01.
The International Classification of Diseases (ICD)
The International Classification of Diseases (ICD) is the international “standard diagnostic tool for epidemiology, health management, and clinical purposes.” Its full official name is International Statistical Classification of Diseases and Related Health Problems.
The ICD is maintained by the World Health Organization (WHO), the directing and coordinating authority for health within the United Nations System. The ICD is designed as a health care classification system, providing a system of diagnostic codes for classifying diseases, including nuanced classifications of a wide variety of signs, symptoms, abnormal findings, complaints, social circumstances, and external causes of injury or disease. This system is designed to map health conditions to corresponding generic categories together with specific variations, assigning them a designated code, up to six characters long. Thus, major categories are designed to include a set of similar diseases.
MeaningCloud shows the codes corresponding to the ICD-10 classification system. It includes more than 155,000 different codes and lets you track many new diagnoses and procedures, a significant expansion on the 17,000 codes that were available in ICD-9.
See the free demo: https://www.meaningcloud.com/demos/health-text-analytics-demo
[1] Wester K, Jönsson AK, Spigset O, Druid H, Staffan H. Incidence of fatal adverse drug reactions: a population based study. Brit J Clin Pharmaco. 2008;65(4):573–579. doi: 10.1111/j.1365-2125.2007.03064.x. [PMC free article] [PubMed] [Cross Ref]
[2] Bond CA, Raehl CL. Adverse drug reactions in United States hospitals. Pharmacotherapy. 2006;26(5):601–608. doi: 10.1592/phco.26.5.601. [PubMed] [Cross Ref]
[3] van Der Hooft CS, Sturkenboom MCJM, van Grootheest K, Kingma HJ, Stricker BHCh. Adverse drug reaction-related hospitalisations. Drug Saf. 2006;29(2):161–168. doi: 10.2165/00002018-200629020-00006. [PubMed] [Cross Ref]


